AUDIT, Analysis & evalUation Dashboard of artIficial inTelligence, is a tool designed to provide researchers and developers with an interactive way to better analyze and explore MRI datasets and segmentation models. Given its functionalities to extract the most relevant features and metrics from your multiple data sources, it allows for uncovering biases both intra and inter-dataset as well as in the model predictions.
Details of our work are provided in our paper AUDIT: An open-source Python library for AI model evaluation with use cases in MRI brain tumor segmentation. We hope that users will leverage AUDIT to gain novel insights into the field of medical image segmentation.
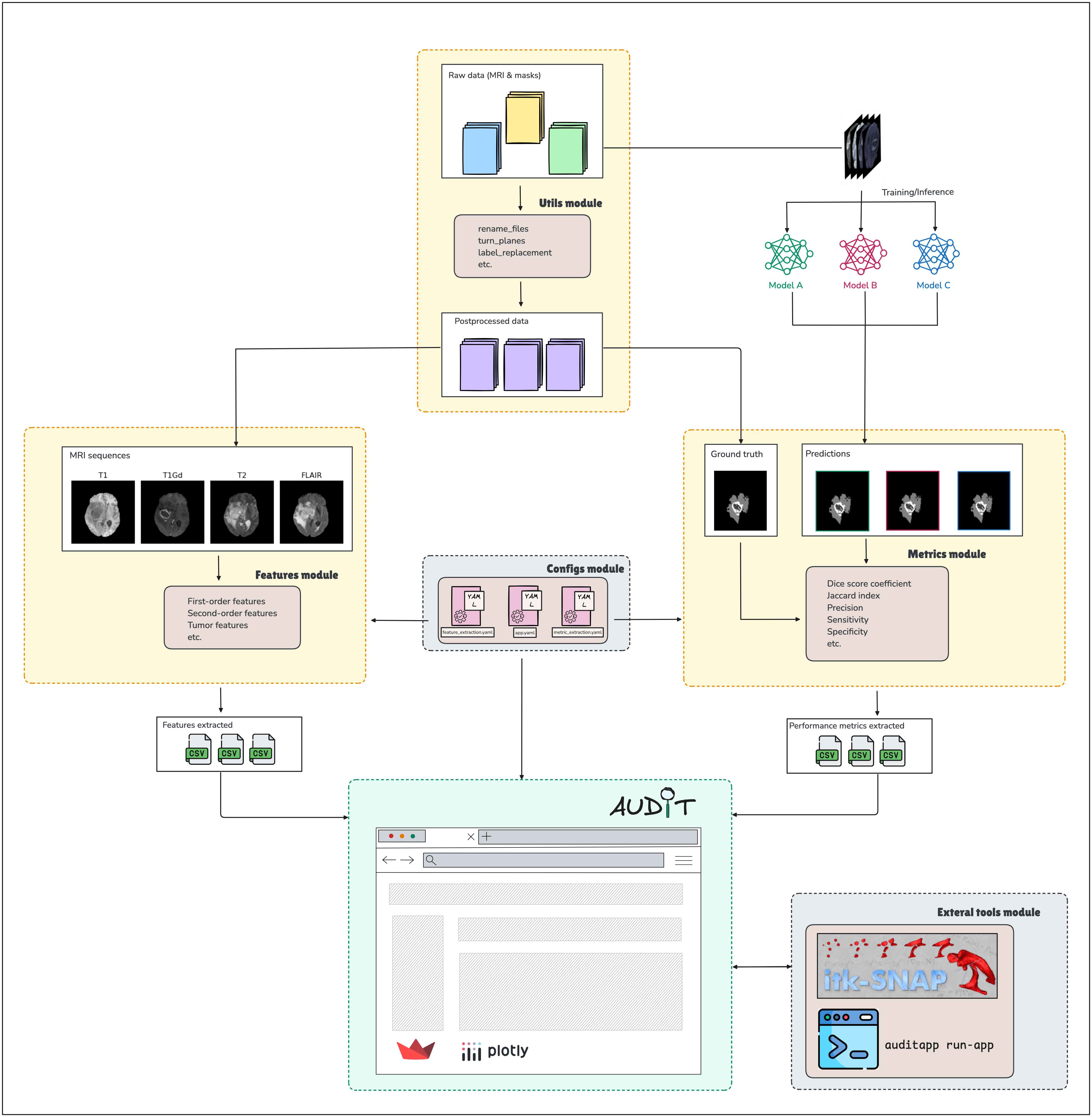
The diagram below illustrates the overall workflow of AUDIT, from input data to data visualization on the APP.
For more details, please refer to the AUDIT paper.
- Home Page: The main landing page of the tool.
- Univariate: Exploration of individual features to understand how they are distributed.
- Multivariate: Analysis of multiple features simultaneously to explore relationships and hidden patterns.
- Segmentation error matrix: Overview of disagreements between ground truth and predicted segmentation through a class-wise error matrix.
- Single model performance: Evaluation of the performance of a single model based on extracted features.
- Pairwise model performance: Perform pairwise comparisons between models to find statistically significant differences.
- Multi-model performance: Comparative analysis of performance metrics across multiple models.
- Longitudinal measurements: Analysis of data collected over time to observe trends and changes in model performance.
- Subjects' exploration: Detailed examination of individual subjects within the dataset.
Last released version of AUDIT is hosted at https://auditapp.streamlitapp.com for an online overview of its functionalities.
AUDIT can be installed either from our GitHub repository or from PyPI using the command pip install auditapp. In this guide, we will show how to install it from the GitHub repository. For a more detailed exploration of AUDIT, please check our official documentation.
Create an isolated Anaconda environment (recommended to avoid dependency conflicts):
conda create -n audit_env python=3.10
conda activate audit_envClone the repository:
git clone https://github.com/caumente/AUDIT.git
cd AUDITInstall the required packages:
pip install -r requirements.txtEdit the config files in ./src/audit/configs/ directory to set up the paths for data loading and other configurations:
This configuration file is used to set paths, labels, features, and longitudinal study parameters for feature extraction in AUDIT.
Show configuration
# Paths to all the datasets
data_paths:
BraTS2020: '/home/usr/AUDIT/datasets/BraTS2020/BraTS2020_images'
BraTS2024_PED: '/home/usr/AUDIT/datasets/BraTS2024_PED/BraTS2024_PED_images'
BraTS2024_SSA: '/home/usr/AUDIT/datasets/BraTS2024_SSA/BraTS2024_SSA_images'
UCSF: '/home/usr/AUDIT/datasets/UCSF/UCSF_images'
LUMIERE: '/home/usr/AUDIT/datasets/LUMIERE/LUMIERE_images'
# Sequences available
sequences:
- '_t1'
- '_t2'
- '_t1ce'
- '_flair'
# Mapping of labels to their numeric values
labels:
BKG: 0
EDE: 3
ENH: 1
NEC: 2
# List of features to extract
features:
statistical: true
texture: true
spatial: true
tumor: true
# Longitudinal study settings
longitudinal:
UCSF:
pattern: "_" # Pattern used for splitting filename
longitudinal_id: 1 # Index position for the subject ID after splitting the filename. Starting by 0
time_point: 2 # Index position for the time point after splitting the filename. Starting by 0
LUMIERE:
pattern: "-"
longitudinal_id: 1
time_point: 3
# Path where extracted features will be saved
output_path: '/home/usr/AUDIT/outputs/features'
logs_path: '/home/usr/AUDIT/logs/features'
# others
cpu_cores: 8This configuration file defines the paths to datasets and model predictions, the labels, metrics to compute, and output paths for metric extraction in AUDIT.
Show configuration
# Path to the raw dataset
data_path: '/home/usr/AUDIT/datasets/BraTS2024_PED/BraTS2024_PED_images'
# Paths to model predictions
model_predictions_paths:
nnUnet: '/home/usr/AUDIT/datasets/BraTS2024_PED/BraTS2024_PED_seg/nnUnet'
SegResNet: '/home/usr/AUDIT/datasets/BraTS2024_PED/BraTS2024_PED_seg/SegResNet'
# Mapping of labels to their numeric values
labels:
BKG: 0
EDE: 3
ENH: 1
NEC: 2
# List of metrics to compute
metrics:
dice: true
jacc: true
accu: true
prec: true
sens: true
spec: true
haus: true
size: true
# Library used for computing all the metrics
package: audit
# Path where output metrics will be saved
output_path: '/home/usr/AUDIT/outputs/metrics'
filename: 'BraTS2024_PED'
logs_path: '/home/usr/AUDIT/logs/metric'
# others
cpu_cores: 8This configuration file sets up paths for datasets, extracted features, metrics, and model predictions. It also defines available sequences and label mappings used by the AUDIT application.
Show configuration
# Sequences available. First of them will be used to compute properties like spacing
sequences:
- '_t1'
- '_t2'
- '_t1ce'
- '_flair'
# Mapping of labels to their numeric values
labels:
BKG: 0
EDE: 3
ENH: 1
NEC: 2
# Root path for datasets, features extracted, and metrics extracted
datasets_path: './datasets' # '/home/usr/AUDIT/datasets'
features_path: './outputs/features' # '/home/usr/AUDIT/outputs/features'
metrics_path: './outputs/metrics' # '/home/usr/AUDIT/outputs/metrics'
# Paths for raw datasets
raw_datasets:
BraTS2020: "${datasets_path}/BraTS2020/BraTS2020_images"
BraTS2024_SSA: "${datasets_path}/BraTS2024_SSA/BraTS2024_SSA_images"
BraTS2024_PED: "${datasets_path}/BraTS2024_PED/BraTS2024_PED_images"
UCSF: "${datasets_path}/UCSF/UCSF_images"
LUMIERE: "${datasets_path}/LUMIERE/LUMIERE_images"
# Paths for feature extraction CSV files
features:
BraTS2020: "${features_path}/extracted_information_BraTS2020.csv"
BraTS2024_SSA: "${features_path}/extracted_information_BraTS2024_SSA.csv"
BraTS2024_PED: "${features_path}/extracted_information_BraTS2024_PED.csv"
UCSF: "${features_path}/extracted_information_UCSF.csv"
LUMIERE: "${features_path}/extracted_information_LUMIERE.csv"
# Paths for metric extraction CSV files
metrics:
BraTS2024_SSA: "${metrics_path}/extracted_information_BraTS2024_SSA.csv"
BraTS2024_PED: "${metrics_path}/extracted_information_BraTS2024_PED.csv"
UCSF: "${metrics_path}/extracted_information_UCSF.csv"
LUMIERE: "${metrics_path}/extracted_information_LUMIERE.csv"
# Paths for models predictions
predictions:
BraTS2024_SSA:
nnUnet: "${datasets_path}/BraTS2024_SSA/BraTS2024_SSA_seg/nnUnet"
SegResNet: "${datasets_path}/BraTS2024_SSA/BraTS2024_SSA_seg/SegResNet"
BraTS2024_PED:
nnUnet: "${datasets_path}/BraTS2024_PED/BraTS2024_PED_seg/nnUnet"
SegResNet: "${datasets_path}/BraTS2024_PED/BraTS2024_PED_seg/SegResNet"
Use the following commands in your terminal to run the Feature extraction and Metric extraction scripts:
python src/audit/feature_extraction.pypython src/audit/metric_extraction.pyAfter running either script, a logs folder will be created to keep track of the execution. All output files will be stored in the folder defined in the corresponding config file (by default, in the outputs folder).
AUDIT app is built on top of the Streamlit library. Use the following command to run the app and start exploring your data:
python src/audit/app/launcher.pyAUDIT can be adjusted for opening cases with ITK-Snap while exploring the data in the different dashboards. The ITK-Snap tool must have been installed and preconfigured before. Below are placeholders for the necessary configuration for each operating system. Information will be added soon.
show configuration
Show configuration
A brief tutorial to install ITK-SNAP 4.4
- Download ITK-SNAP 4.4
wget https://downloads.itksnap.org/itksnap-4.4.0-20250909-Linux-x86_64.tar.gz- Extract and move to
/opt
sudo tar -xvzf itksnap-4.4.0-20250909-Linux-x86_64.tar.gz -C /opt
sudo mv /opt/itksnap-4.4.0-20250909-Linux-x86_64 /opt/itksnap- Create a wrapper script to run from anywhere
sudo nano /usr/local/bin/itksnapFile content:
#!/bin/bash
DIR=/opt/itksnap
"$DIR/bin/itksnap" "$@"Make it executable:
sudo chmod +x /usr/local/bin/itksnap- Run ITK-SNAP 4.4
itksnapPlease feel free to contact us with any issues, comments, or questions.
- Carlos Aumente (UO297103@uniovi.es)
- Mauricio Reyes
- Michael Muller
- Jorge Díez
- Beatriz Remeseiro
Apache License 2.0