-
Notifications
You must be signed in to change notification settings - Fork 6
Home
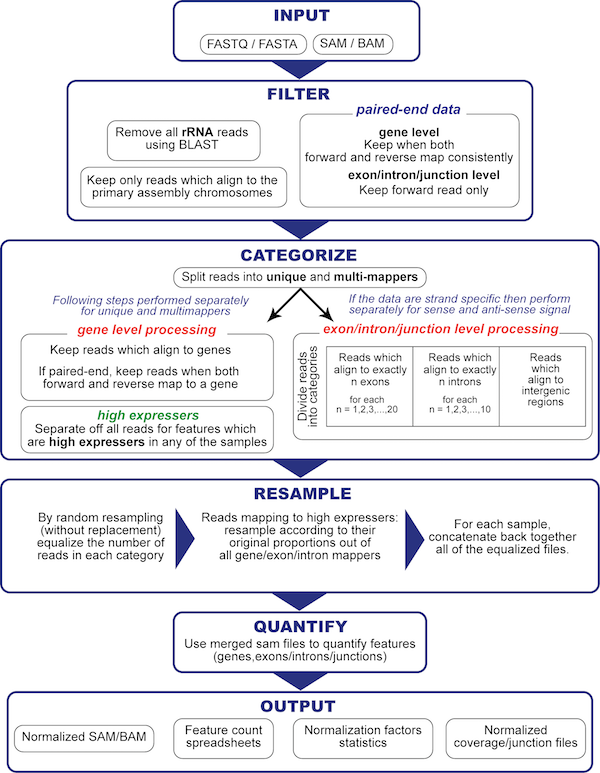
PORT is a resampling based read-level normalization and quantification pipeline for RNA-Seq differential expression analysis.
Unlike most existing RNA-Seq processing pipelines, PORT normalizes first and performs quantification on the normalized reads. PORT’s normalization approach addresses each identified normalization factor systematically, rather than treating normalization as a global problem that can be dealt with as a single factor. Here’s a list of normalization factors PORT address that vary between samples:
- Total read count
- Ribosomal content
- Fragment length
- Mitochondrial content
- Proportion of multi-mappers
- Sense vs. anti-sense transcription
- Very highly expressed and variable features*
- Proportion of gene-mappers (Gene level normalization)
- Percent exon to non-exon signal (Exon-Intron-Junction level normalization)
- Proportion of intergenic to intronic signal (Exon-Intron-Junction level normalization)
- Proportion of one-exon/intron to multi-exon/intron mappers (Exon-Intron-Junction level normalization)
* features: gene, exon, introns
PORT offers two types of normalization: GENE and EXON-INTRON-JUNCTION level normalization. It uses resampling to equalize different categories of reads across all samples independently and then merges them together into a final set of reads.
PORT Flowchart
next: Getting started